Background Theory on The Outlier Detection Model
This vignette is an introduction to the “behind-the-scenes” in the paper Lindskou, Svante Eriksen, and Tvedebrink (2019)
Contingency Tables
The outlier detection model can only be used on discrete data; i.e data for which all variables can only take on a finite set of values (also called levels). We also say, that such variables are discrete variables or categorical variables.
Consider for example the built in discrete data set in R on 4526 applicants to graduate school at Berkeley for the six largest departments in 1973 classified by admission and sex. Below, a small sample of the data is seen.
| Admit | Gender | Dept |
|---|---|---|
| Rejected | Female | D |
| Admitted | Male | B |
| Rejected | Female | C |
| Rejected | Male | E |
| Rejected | Female | F |
| Rejected | Female | C |
Another and more appropriate way of showing the entire data, is via a contingency table which summarizes the counts of all combinations of Admit, Gender and Dept
| Dept | A | B | C | D | E | F | ||
| Admit | Gender | |||||||
| Admitted | Female | 89 | 17 | 202 | 131 | 94 | 24 | |
| Male | 512 | 353 | 120 | 138 | 53 | 22 | ||
| Rejected | Female | 19 | 8 | 391 | 244 | 299 | 317 | |
| Male | 313 | 207 | 205 | 279 | 138 | 351 |
This is much more informative and we can e.g. say immediately that 207 Male applicants were Rejected at department B. A particular entry in the table is called a cell. So (Male, Rejected, B) is a cell with cellcount 207. The dimension of the table is the number of variables (here 3). For high-dimensional tables things get more complicated and it may not be feasible to show the table (think of hundreds of DNA markers) and there may be many cells with zero cellcount.
The outlier detection method (described shortly) uses such contingency table data. However, it should be mentioned one more time, that a contingency table is in one-to-one correspondence with a “regular” categorical data set; contingency tables are nothing more than a convenient way of describing categorical data sets.
Outliers and Novelties
An outlier can be regarded as an observation which deviates so much from the other observations in a database as to arouse suspicions that it was generated by a different mechanism. The outlier detection method directly adapts this definition by specifying a hypothesis of an outlier being distributed differently than all other observations in a given database. An outlier is also a very case-specific unit, since it may be interpreted as natural extreme noise in some applications and in other applications it might be the most interesting observation.
Outlier detection is usually divided into (yes the same word) where the task is to detect observations that are extreme within some dataset. In combination to categorical data this amounts to detect unusual combinations between all variables in consideration. On the other hand, we might be interested in detecting if a new (novel) observation is an outlier in a homogeneous dataset which is sometimes referred to as in the litterature. molic handles both of these cases through a single function called fit_outlier.
For high-dimensional data, it is a tedious task to determine anomalies such as outliers and novelties. The method described here is a multivariate method for outlier detection in high-dimensional contingency tables, i.e. data with discrete variables only.
The method relies on the class of decomposable graphical models to model the relationship among the variables of interest, which can be depicted by an undirected graph called the interaction graph. molic requires the ess package to be installed. The ess package offers the simple fit_graph function that conveniently fits a decomposable graph to data.
Decomposable Graphical Models
In order to model the interaction structure between variables, we need to construct a decomposable graphical model. A graphical model is a statistical model for which an undirected graph represents the interaction between the vertices in the model. An undirected graph is a pair \(G = (V,E)\) where \(V\) is a set of vertices and \(E\) is a set of edges connecting elements in \(V\). An edge connecting two vertices indicates that these two are dependend on each other; this is also called a two-way interaction. A threeway interaction occurs when three vertices are all mutually connected (e.g. can be visualized as a triangle). An undirected graph is decomposable if there are no cycles of length greater than four without a chord (an edge between two non-adjacent vertices in the cycle). The subgraph \(G_{A} = (A, E_{A})\) consist of vertices \(A\subseteq V\) from \(G\) and the corresponding edges \(E_A\) between them. A graph is complete if there is an edge between all pairs of vertices and a complete subgraph is called a (maximal) clique if it is not contained in any other complete subgraph. A subset of vertices is complete if it induces a complete subgraph. Two sets \(A,B \subseteq V\) are separated by a third set \(C \subseteq V\) if all paths between vertices in \(A\) and \(B\) go through \(C\). If \(C\) is the smallest set such that \(A\) and \(B\) are separated, we say that \(C\) is a (minimal) separator for \(A\) and \(B\).
Consider the undirected graph \(G\) in the figure below.
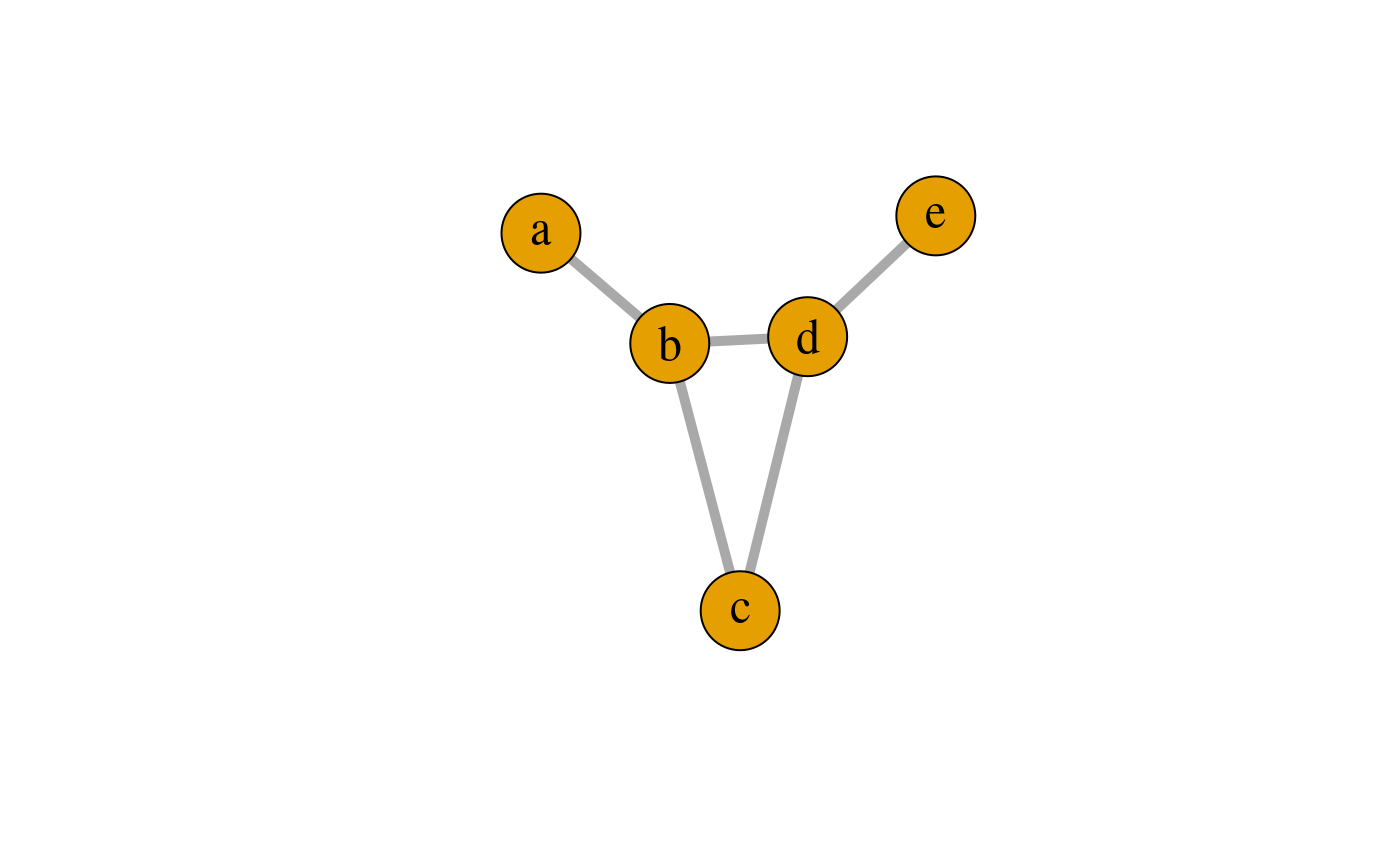
\(G\): An undirected decomposable graph.
The set of vertices is \(V = \{a,b,c,d,e\}\) and the set of edges is \(E = \{ab, bc, bd, cd,de\}\). The cliques are \(C_1 = \{a, b\}\), \(C_2 = \{b, c, d\}\) and \(C_3 = \{d, e\}\). The minimal separators are \(S_{2} = \{b\}\) and \(S_3 = \{d\}\) where \(S_{2}\) separates \(C_{1}\) and \(C_{2}\) and \(S_{3}\) separates \(C_{2}\) and \(C_{3}\). Since \(G\) has no cycles of length greater than three, the graph is decomposable. Notice, that \(a\) is only connected to \(\{c,d,e\}\) through \(\{b\}\); we interpret this as “\(a\) is independent of \(\{c,d,e\}\) when we know the value of \(b\)”. Such statements can be used to gain insight into complex structures.
Finally, we can associate a probability measure with an interaction graph; an undirected graph with each vertex being a random variable. For decomposable graphs, the probability density function can be written in terms of the cliques and separators. Collectively, models for which the interaction graph is decomposable are called decomposable graphical models (DGM). For more details on graphical models, see for example (Whittaker 2009; Lauritzen 1996).
Tree graphs are per definition decomposable graphs. Notice that the graph \(G\) is not a tree because of the presence of the cycle \(\{b,c,d\}\). However, the graph is indeed decomposable.
The Test Statistic
Assume that the likelihood \(L_0\) expresses how likely it is that \(z_{new}\) belongs to the database \(D\). We can also specify an alternative likelihood, \(L_{1}\), specifying how likely it is that \(z_{new}\) do not belong to \(D\). We then define the likelihood ratio as \[\begin{equation*} \label{eq:LR} LR = \frac{L_{0}}{L_{1}}, \end{equation*}\]which can be shown to be completely specified through the counts of observations in cliques and separators for the given interaction graph. We can therefore test if \(z_{new}\) is an outlier in \(D\) by calculating \(LR\) and determine if the value of \(LR\) is “too large” in which case we would reject that \(z_{new}\) comes from \(D\).
A note on novelty detection
It should be noted, that the likelihood ratio defined above, applies to novelty detection; i.e. we test if the novel observation \(z_{new}\) is an outlier in \(D\). If the goal is to find the outliers in \(D\) one should, one at a time, remove the observations from \(D\) and regard the observations as novel observations and apply the above test statistic. All of this is taken care of in molic via the fit_outlier function.
References
Lauritzen, Steffen L. 1996. Graphical Models. Vol. 17. Clarendon Press.
Lindskou, Mads, Poul Svante Eriksen, and Torben Tvedebrink. 2019. “Outlier Detection in Contingency Tables Using Decomposable Graphical Models.” Scandinavian Journal of Statistics. Wiley Online Library. doi:10.1111/sjos.12407.
Whittaker, Joe. 2009. Graphical Models in Applied Multivariate Statistics. Wiley Publishing.