Conduct multiple novelty tests for a new observation
fit_multiple_models( A, z, response, alpha = 0.05, type = "fwd", q = 0.5, comp = NULL, nsim = 10000, ncores = 1, trace = TRUE, validate = TRUE )
Arguments
| A | A character matrix or data.frame |
|---|---|
| z | Named vector (same names as |
| response | A character with the name of the class variable of interest |
| alpha | The significance level |
| type | Character ("fwd", "bwd", "tree" or "tfwd") - the type of interaction graph to be used |
| q | Penalty term in the stopping criterion when fitting the interaction graph ( |
| comp | A list with character vectors. Each element in the list is a component in the graph (using expert knowledge) |
| nsim | Number of simulations |
| ncores | Number of cores to use in parallelization |
| trace | Logical indicating whether or not to trace the procedure |
| validate | Logical. If true, it checks if |
Value
An object of type multiple_models; a list of of novely objects from which one
can query pvalues etc. for outlierdetection.
See also
Examples
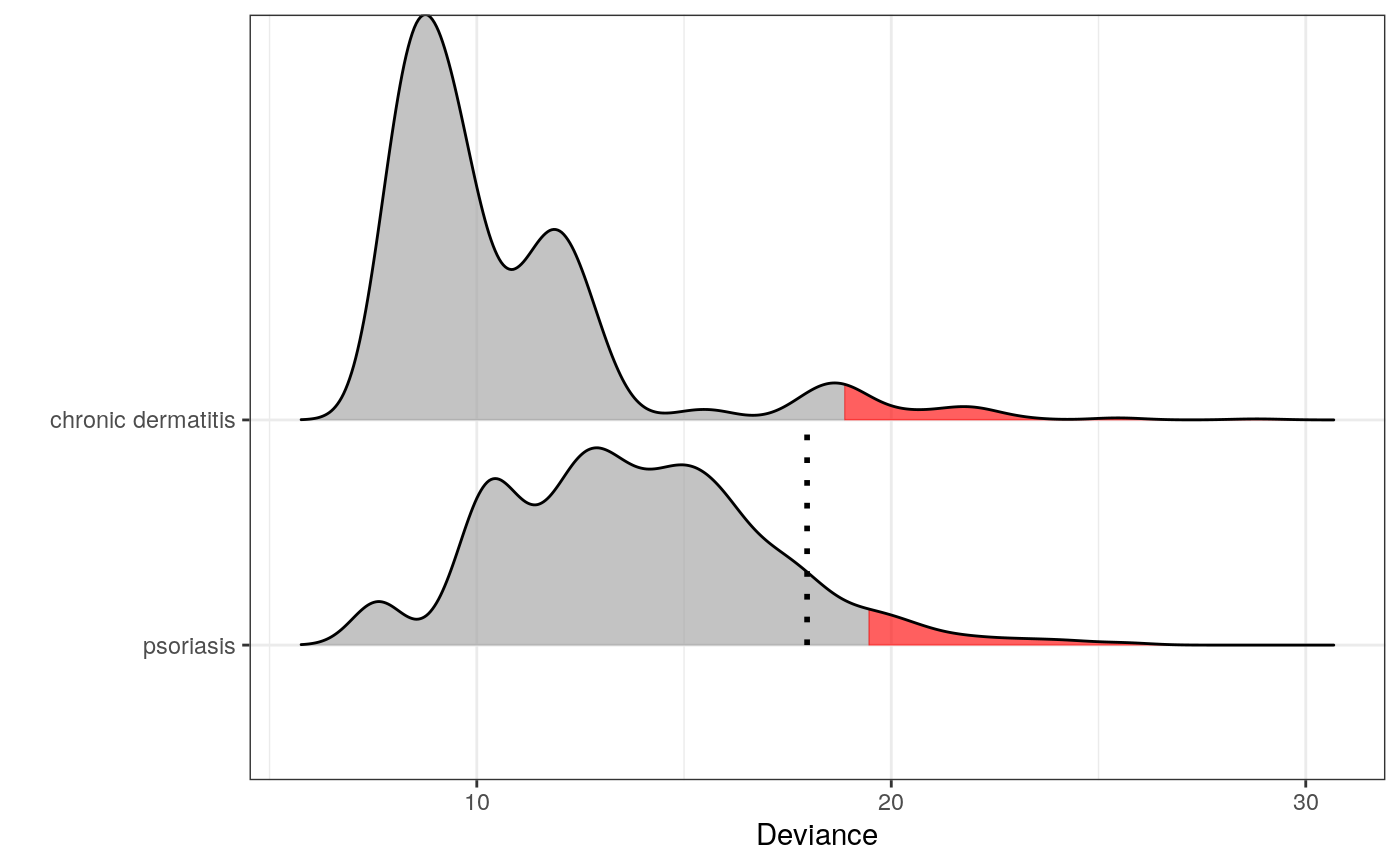
library(dplyr) set.seed(1) # A patient with psoriasis z <- unlist(derma[2, 1:10]) d <- derma[, c(names(z), "ES")] %>% filter(ES %in% c("chronic dermatitis", "psoriasis")) m <- fit_multiple_models(d, z, "ES", nsim = 1000, trace = FALSE, validate = FALSE) plot(m)#>print(m)#> $psoriasis #> #> -------------------------------- #> Simulations: 1000 #> Variables: 10 #> Observations: 112 #> Estimated mean: 13.79 #> Estimated variance: 10.17 #> -------------------------------- #> Critical value: 19.44595 #> Deviance: 17.96901 #> P-value: 0.092 #> Alpha: 0.05 #> <novelty, outlier_model, list> #> -------------------------------- #> #> $`chronic dermatitis` #> #> -------------------------------- #> Simulations: 1000 #> Variables: 10 #> Observations: 49 #> Estimated mean: 10.61 #> Estimated variance: 9.64 #> -------------------------------- #> Critical value: 18.83373 #> Deviance: 34.74245 #> P-value: 0 #> Alpha: 0.05 #> <novelty, outlier_model, list> #> -------------------------------- #> #> attr(,"class") #> [1] "multiple_models" "list"