A function for outlier detection with mixed, but independen, information
fit_mixed_outlier(m1, m2)
Arguments
| m1 | An object returned from |
|---|---|
| m2 | An object returned from |
Value
An object of type mixed_outlier with novelty or outlier
as child classes. These are used for different purposes. See fit_outlier.
Details
It is assumed that the input data to m1 and m2
holds information about the same observation in corresponding rows.
Thus, the two datasets must also be of same dimension.
See also
Examples
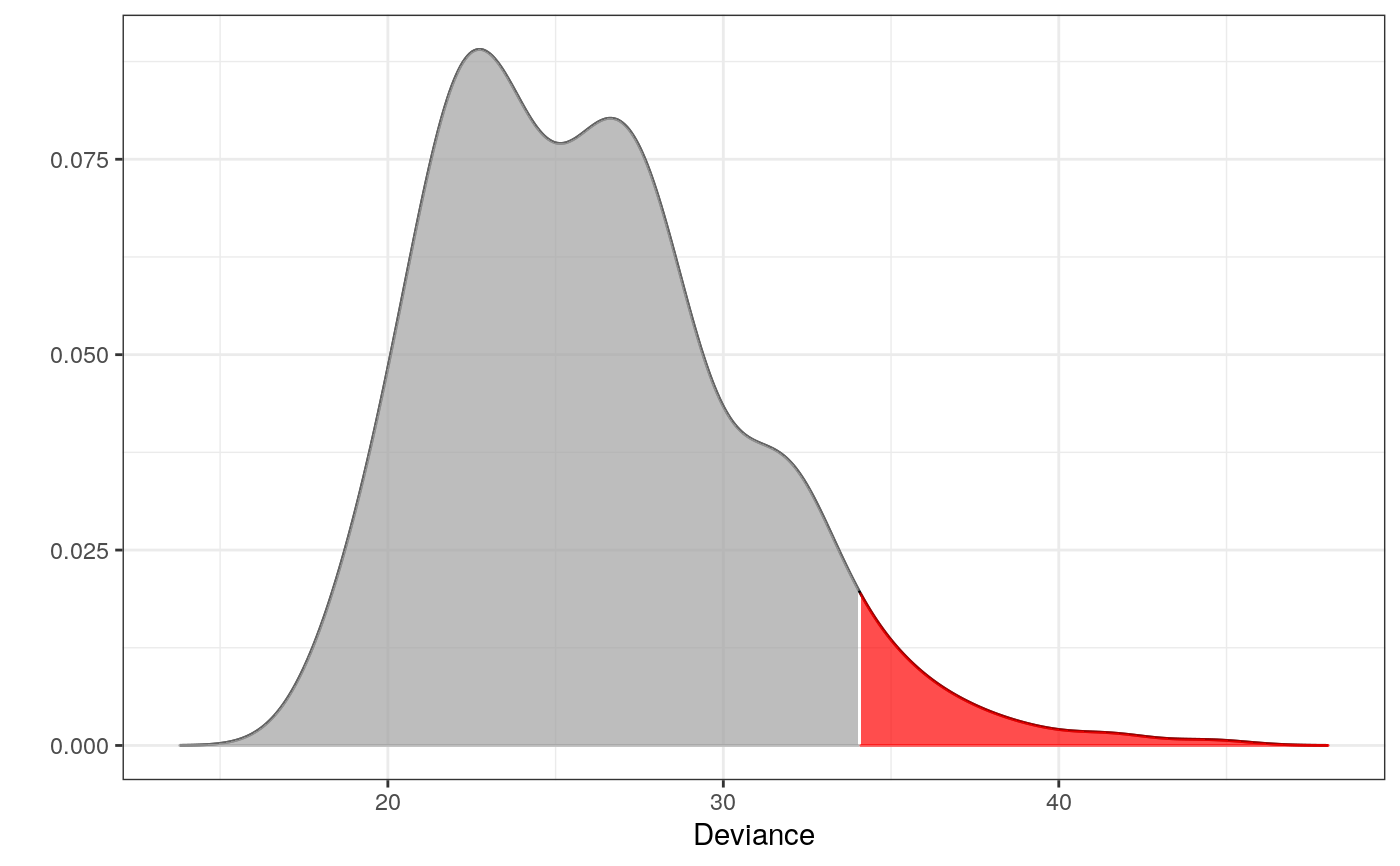
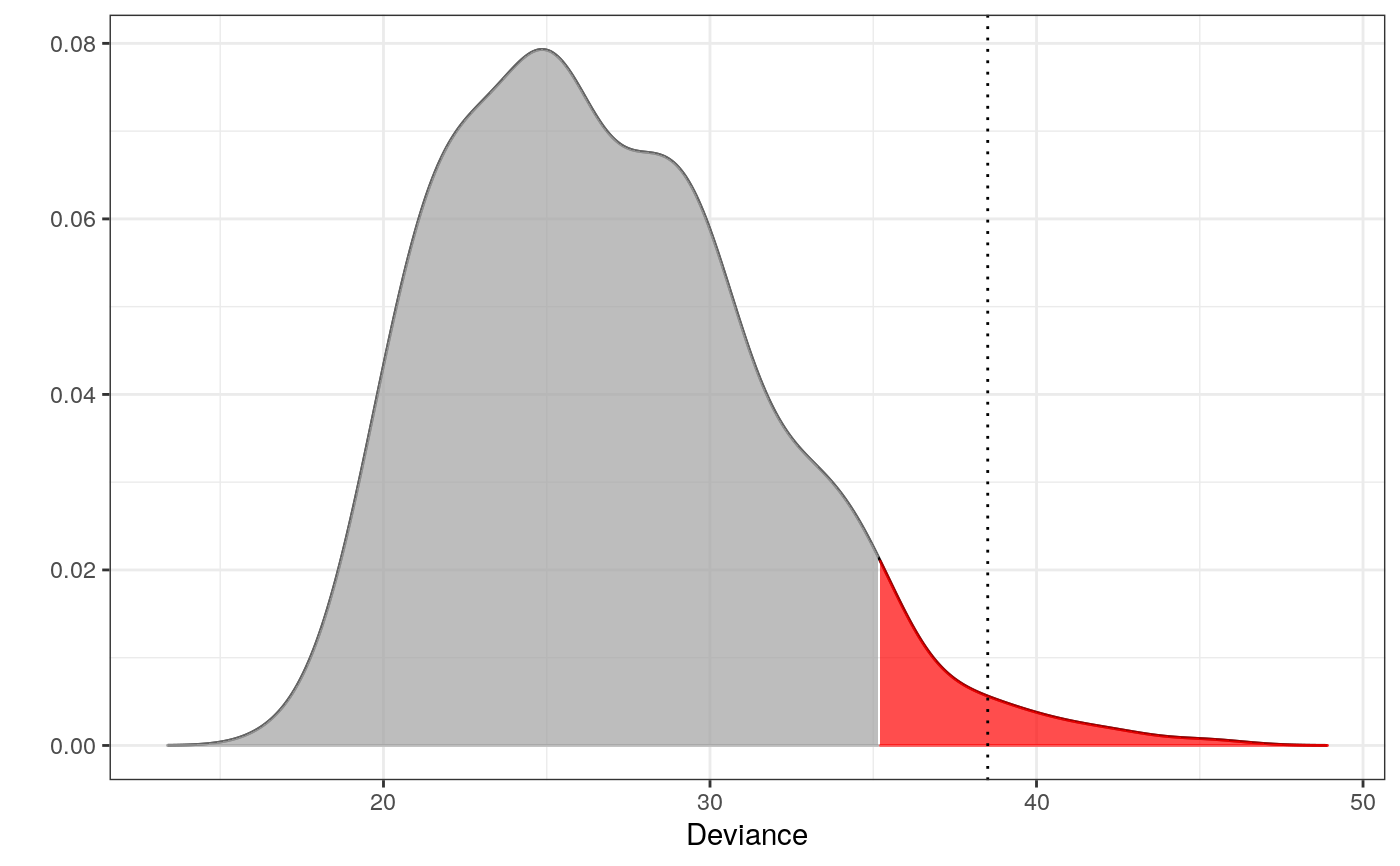
library(dplyr)#> #>#> #> #>#> #> #>#> #> #>set.seed(7) # for reproducibility ## Data # The components - here microhaplotypes haps <- tgp_haps[1:5] # only a subset of data is used to exemplify dat <- tgp_dat %>% select(pop_meta, sample_name, all_of(unname(unlist(haps)))) # All the Europeans eur <- dat %>% as_tibble() %>% filter(pop_meta == "EUR") # Extracting the two databases for each copy of the chromosomes eur_a <- eur %>% filter(grepl("a$", sample_name)) %>% select(-c(1:2)) eur_b <- eur %>% filter(grepl("b$", sample_name)) %>% select(-c(1:2)) # Fitting the interaction graphs on the EUR data ga <- fit_components(eur_a, comp = haps, trace = FALSE) gb <- fit_components(eur_b, comp = haps, trace = FALSE) print(ga)#> A Decomposable Graph With #> ------------------------- #> Nodes: 17 #> Edges: 18 / 136 #> Cliques: 7 #> - max: 4 #> - min: 1 #> - avg: 2.57 #> <gengraph, list> #> -------------------------## --------------------------------------------------------- ## EXAMPLE 1 ## Testing which observations within data are outliers ## --------------------------------------------------------- # Only 500 simulations is used here to exeplify # The default number of simulations is 10,000 m1 <- fit_outlier(eur_a, ga, nsim = 500) # consider using more cores (ncores argument) m2 <- fit_outlier(eur_b, gb, nsim = 500) # consider using more cores (ncores argument) m <- fit_mixed_outlier(m1, m2) print(m)#> #> -------------------------------- #> Simulations: 500 #> Variables: 17 #> Observations: 503 #> Estimated mean: 25.95 #> Estimated variance: 20.47 #> -------------------------------- #> Critical value: 34.03098 #> Alpha: 0.05 #> <mixed_outlier, outlier, outlier_model, list> #> --------------------------------plot(m)outs <- outliers(m) eur_a_outs <- eur_a[which(outs), ] eur_b_outs <- eur_b[which(outs), ] # Retrieving the test statistic for individual observations x1 <- rbind(eur_a_outs[1, ], eur_b_outs[1, ]) x2 <- rbind(eur_a[1, ], eur_b[1, ]) dev1 <- deviance(m, x1) # falls within the critical region in the plot (the red area) dev2 <- deviance(m, x2) # falls within the acceptable region in the plot dev1#> [1] 39.01466dev2#> [1] 27.03032#> [1] 0.008#> [1] 0.372# \donttest{ ## --------------------------------------------------------- ## EXAMPLE 2 ## Testing if a new observation is an outlier ## --------------------------------------------------------- # Testing if an American is an outlier in Europe amr <- dat %>% as_tibble() %>% filter(pop_meta == "AMR") z1 <- amr %>% filter(grepl("a$", sample_name)) %>% select(unname(unlist(haps))) %>% slice(1) %>% unlist() z2 <- amr %>% filter(grepl("b$", sample_name)) %>% select(unname(unlist(haps))) %>% slice(1) %>% unlist() # Only 500 simulations is used here to exemplify # The default number of simulations is 10,000 m3 <- fit_outlier(eur_a, ga, z1, nsim = 500) # consider using more cores (ncores argument) m4 <- fit_outlier(eur_b, gb, z2, nsim = 500) # consider using more cores (ncores argument) m5 <- fit_mixed_outlier(m3, m4) print(m5)#> #> -------------------------------- #> Simulations: 500 #> Variables: 17 #> Observations: 504 #> Estimated mean: 26.74 #> Estimated variance: 22.3 #> -------------------------------- #> Critical value: 35.1841 #> Deviance: 38.50607 #> P-value: 0.016 #> Alpha: 0.05 #> <mixed_outlier, novelty, outlier_model, list> #> --------------------------------plot(m5)# }